learnDBN
A Java implementation for learning Dynamic Bayesian Networks.
Released under the Apache License 2.0
Program description
learnDBN is a Java implementation of a Dynamic Bayesian Network (DBN) structure learning algorithm. It can learn tDBN, cDBN and bcDBN structures from a file with multivariate longitudinal observations. Also, it improves these algorithms by allowing the data to have missing values. As such this implementation can impute missing values.
Current release
This is the first implementation of this program. It comes packaged as an executable JAR file, already including the required external libraries.
Usage
By executing the jar file …
$ java -jar learnDBN.jar
… the available command-line options are shown:
usage: learnDBN
-bcDBN,--bcDBN Learns a bcDBN structure.
-c,--compact Outputs network in compact format, omitting
intra-slice edges. Only works if specified
together with -d and with --markovLag 1.
-cDBN,--cDBN Learns a cDBN structure.
-d,--dotFormat Outputs network in dot format, allowing
direct redirection into Graphviz to
visualize the graph.
-i,--file <file> Input CSV file to be used for network
learning.
-imp,--impute If the file has missing values impute these
values. The resulting data with imputed
values is saved in the same folder with
<filename>_imputed.csv
-ind,--intra_in <int> In-degree of the intra-slice network
-m,--markovLag <int> Maximum Markov lag to be considered, which
is the longest distance between connected
time-slices. Default is 1, allowing edges
from one preceding slice.
-mt,--MultiThread Learns the DBN using parallel computations.
-ns,--nonStationary Learns a non-stationary network (one
transition network per time transition). By
default, a stationary DBN is learnt.
-o,--outputFile <file> Writes output to <file>. If not supplied,
output is written to terminal.
-p,--numParents <int> Maximum number of parents from preceding
time-slice(s).
-pm,--parameters Learns and outputs the network parameters.
-r,--root <int> Root node of the intra-slice tree. By
default, root is arbitrary.
-s,--scoringFunction <arg> Scoring function to be used, either MDL or
LL. MDL is used by default.
-sp,--spanning Forces intra-slice connectivity to be a tree
instead of a forest, eventually producing a
structure with a lower score.
Input file format
The input file should be in comma-separated values (CSV) format.
- The first line is the header, naming the attributes and specifying the time slice index, separared by two underscores: “attributeName__t”
- The order of the attributes must be maintained: “X1__1”, “X2__1”, “X1__2”, “X2__2”.
- The first column contains an identification (string or number) of each subject (this identifier does not affect the learnt network).
- All other lines correspond to observations of an individual over time.
- Missing values can be marked as “?” but should not occur, as the algorithm discards the observation (time slice) in question.
- The variables can have numerical and categorical values.
A very simplistic input file example is the following:
"subject_id","X1__0","X2__0","X3__0","X1__1","X2__1","X3__1","X1__2","X2__2","X3__2"
"6","7.0","40.0","5.0","7.0","20.0","5.0","4.0","20.0","5.0"
"7","4.0","40.0","5.0","7.0","40.0","5.0","7.0","40.0","5.0"
"8","7.0","20.0","5.0","7.0","40.0","5.0","4.0","20.0","9.0"
"9","7.0","40.0","9.0","7.0","20.0","5.0","7.0","40.0","?"
"10","7.0","20.0","5.0","4.0","20.0","9.0","7.0","20.0","9.0"
"11","?","20.0","5.0","?","20.0","5.0","4.0","20.0","9.0"
"12","4.0","20.0","5.0","7.0","20.0","5.0","4.0","20.0","9.0"
Example
The first example considers a synthetic network structure with 5 attributes, each one taking 4 states and one parent from the preceding slice ([t] denotes the time slice):
The above network was sample to produce the following file:
- exmp1.csv, with 1000 observations with 5 time steps, 20% of missing values, 20% of observations with missing values
As all nodes have exactly one parent from the past, the best options are to limit the number of parents with -p 1.
The command to learn the network and impute the missing values is:
java -jar learnDBN.jar -i exmp1.csv -p 1 -imp -mt
which produces the following output:
Evaluating network with MDL score.
Found missing values in data.
Parameter EM step: 1 score: -32184.7968939429
Parameter EM step: 2 score: -32116.81895576152
Parameter EM step: 3 score: -32115.385956588492
Parameter EM step: 4 score: -32115.342586060127
Parameter EM step: 5 score: -32115.34083770815
Parameter EM step: 6 score: -32115.340752037384
Parameter EM step: 7 score: -32115.34074725887
Parameter EM step: 8 score: -32115.3407469682
Parameter EM step: 9 score: -32115.340746949463
Parameter EM step: 10 score: -32115.3407469482
Parameter EM step: 11 score: -32115.340746948117
Parameter EM step: 12 score: -32115.340746948106
Parameter EM step: 13 score: -32115.340746948103
Parameter EM step: 14 score: -32115.340746948103
Strutural EM step: 1
Parameter EM step: 1 score: -27336.936378385602
Parameter EM step: 2 score: -27015.449042522705
Parameter EM step: 3 score: -27009.25438525708
Parameter EM step: 4 score: -27009.088509908135
Parameter EM step: 5 score: -27009.082936546387
Parameter EM step: 6 score: -27009.082723151798
Parameter EM step: 7 score: -27009.08271432131
Parameter EM step: 8 score: -27009.082713937845
Parameter EM step: 9 score: -27009.082713920747
Parameter EM step: 10 score: -27009.082713919976
Parameter EM step: 11 score: -27009.082713919954
Parameter EM step: 12 score: -27009.082713919946
Parameter EM step: 13 score: -27009.082713919946
Strutural EM step: 2
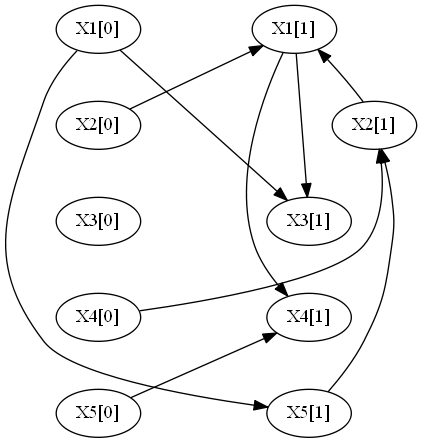
-----------------
X2[0] -> X1[1]
X4[0] -> X2[1]
X1[0] -> X3[1]
X5[0] -> X4[1]
X1[0] -> X5[1]
X2[1] -> X1[1]
X5[1] -> X2[1]
X1[1] -> X3[1]
X1[1] -> X4[1]
Activating the -d switch to directly output in dot format, in order to this functionality to work you need to install Graphviz in the following directory:
- Windows: C:\Program Files (x86)\Graphviz2.38
- Mac: /usr/local/bin/dot
- Linux: /usr/bin/dot
However you can change this directory by editing the GraphViz.java file and then compile the program.
So running the following command: ´´´shell java -jar learnDBN.jar -i exmp1.csv -p 1 -imp -mt -d ´´´
Produces the imputed file exmp1_imputed and the following graph: