learnDBM
A Java implementation for learning Dynamic Bayesian Multinets.
Released under the Apache License 2.0
Program description
learnDBM is a Java implementation of a Dynamic Bayesian Multinet (DBM) structure learning algorithm. Moreover, by using the DBM model this implementation has the capability of performing clustering on the data.
Current release
This is the first implementation of this program. It comes packaged as an executable JAR file, already including the required external libraries.
Usage
By executing the jar file …
$ java -jar learnDBM.jar
… the available command-line options are shown:
usage: learnDBM
-bcDBN,--bcDBN Learns a bcDBN structure.
-c,--compact Outputs network in compact format, omitting
intra-slice edges. Only works if specified
together with -d and with --markovLag 1.
-cDBN,--cDBN Learns a cDBN structure.
-d,--dotFormat Outputs network in dot format, allowing
direct redirection into Graphviz to
visualize the graph.
-i,--file <file> Input CSV file to be used for network
learning.
-ind,--intra_in <int> In-degree of the intra-slice network
-k,--numClusters <int> Number of cluster in data.
-m,--markovLag <int> Maximum Markov lag to be considered, which
is the longest distance between connected
time-slices. Default is 1, allowing edges
from one preceding slice.
-mt,--MultiThread Learns the DBN using parallel computations.
-ns,--nonStationary Learns a non-stationary network (one
transition network per time transition). By
default, a stationary DBN is learnt.
-o,--outputFile <file> Writes output to <file>. If not supplied,
output is written to terminal.
-p,--numParents <int> Maximum number of parents from preceding
time-slice(s). The default values is 1.
-pm,--parameters Learns and outputs the network parameters.
-sp,--spanning Forces intra-slice connectivity to be a tree
instead of a forest, eventually producing a
structure with a lower score.
Input file format
The input file should be in comma-separated values (CSV) format.
- The first line is the header, naming the attributes and specifying the time slice index, separared by two underscores: “attributeName__t”
- The order of the attributes must be maintained: “X1__1”, “X2__1”, “X1__2”, “X2__2”.
- The first column contains an identification (string or number) of each subject (this identifier does not affect the learnt network).
- All other lines correspond to observations of an individual over time.
- Missing values can be marked as “?” but should not occur, as the algorithm discards the observation (time slice) in question.
- The variables can have numerical and categorical values.
A very simplistic input file example is the following:
"subject_id","X1__0","X2__0","X3__0","X1__1","X2__1","X3__1","X1__2","X2__2","X3__2"
"6","7.0","40.0","5.0","7.0","20.0","5.0","4.0","20.0","5.0"
"7","4.0","40.0","5.0","7.0","40.0","5.0","7.0","40.0","5.0"
"8","7.0","20.0","5.0","7.0","40.0","5.0","4.0","20.0","9.0"
"9","7.0","40.0","9.0","7.0","20.0","5.0","7.0","40.0","?"
"10","7.0","20.0","5.0","4.0","20.0","9.0","7.0","20.0","9.0"
"11","?","20.0","5.0","?","20.0","5.0","4.0","20.0","9.0"
"12","4.0","20.0","5.0","7.0","20.0","5.0","4.0","20.0","9.0"
Example
This example consideres a synthetic dataset generated by 2 DBNs with 5 attributes and 10 time steps.
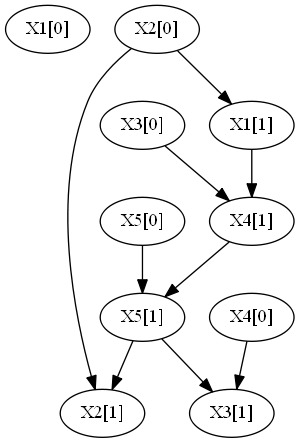
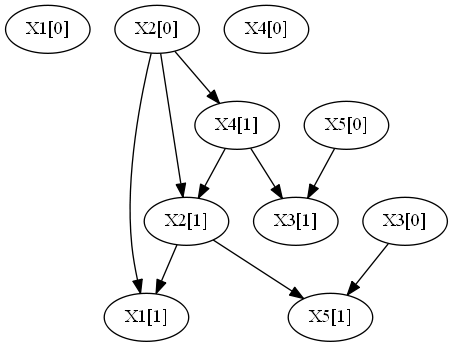
Each of the above networks was sample to produce the following file:
- combinedDataset.csv, with 500 observations with 10 time steps from each DBN
The command to learn the networks and compute the clusters is:
java -jar learnDBM.jar -i ./combinedDataset -k 2 -o /output.csv -mt -d
Which outputs:
Starting with stochastic EM.
Number of clusters : 2
Number of Observations : 2000
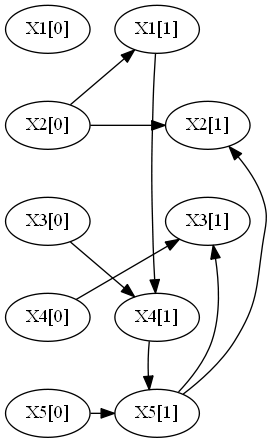
--- Cluster 0 ---
X2[0] -> X1[1]
X2[0] -> X2[1]
X4[0] -> X3[1]
X3[0] -> X4[1]
X5[0] -> X5[1]
X5[1] -> X2[1]
X5[1] -> X3[1]
X1[1] -> X4[1]
X4[1] -> X5[1]
Alpha: 0.5
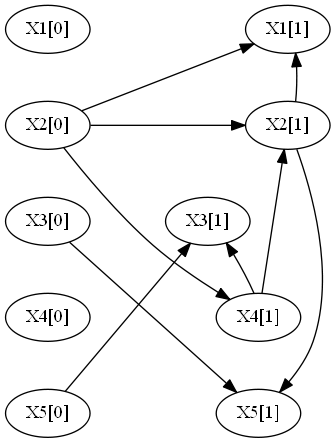
--- Cluster 1 ---
X2[0] -> X1[1]
X2[0] -> X2[1]
X5[0] -> X3[1]
X2[0] -> X4[1]
X3[0] -> X5[1]
X2[1] -> X1[1]
X4[1] -> X2[1]
X4[1] -> X3[1]
X2[1] -> X5[1]
Alpha: 0.5
BIC Score: -96415.86555001826
The flag -d produces the following:
The clustering output has the following Cluster Validity Indexes (CVIs):
| CVI | Score |
|---|---|
| ARI | 0.99800000 |
| RI | 0.9990000 |
| J | 0.9980010 |
| FM | 0.9989995 |
| VI | 0.003434294 |